Help Home > The Download Area
![]()
The Download Area
All JGI portal sites offer an area for downloading primary sequence, annotation, and other data. For older genome assemblies, data is provided through individual download pages which simply list available data files with direct links to download each file. In these cases, please follow any special instructions given on the page.
For newer genomes, and for all Genome Groups, many new file types are now supported: All RAW, Assemblies, Quality Control, various analysis files. All files are accessible in a variety of ways each designed with a particular class of user in mind.
We are setup to allow easy downloads of individual projects/genomes and don't provide means to download whole collections of data. For your convenience and fast downloads you can use Globus and API. Our raw reads are also published to SRA at NCBI for bulk download needs.
NEW. Download with CART
The Genome Portal Cart feature allows you to download data from multiple portals simultaneously.
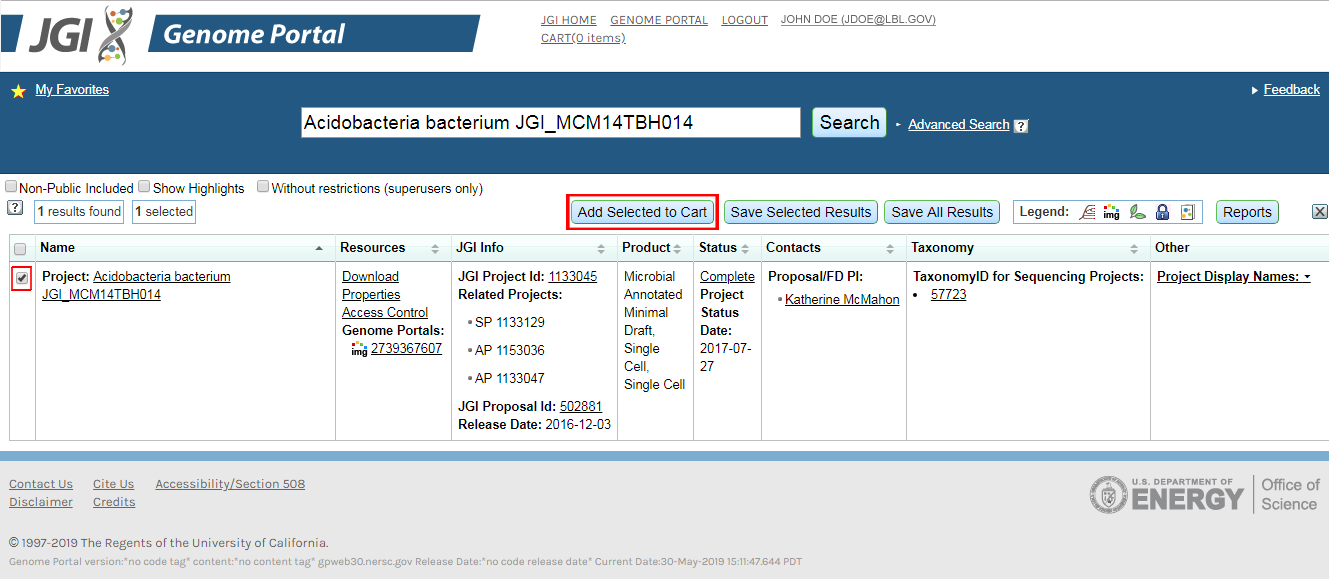
Adding Portals to the Cart (Figure A)
There are a few ways to deposit the data sets of your interest to CART
- From Portal Search Results:
Execute a search. Select the data sets you would like to add to the cart with the checkbox on the left and press "Add selected to Cart" - From "My Favorites":
From your "My Favorites" page, open your group of favorites, add portals to the cart using the "Option 1: From Portal Search Results" instructions - From Proposal/Group Info page:
Go to the Proposal/Group. Select the project(s). Press "Add selected to Cart" button.
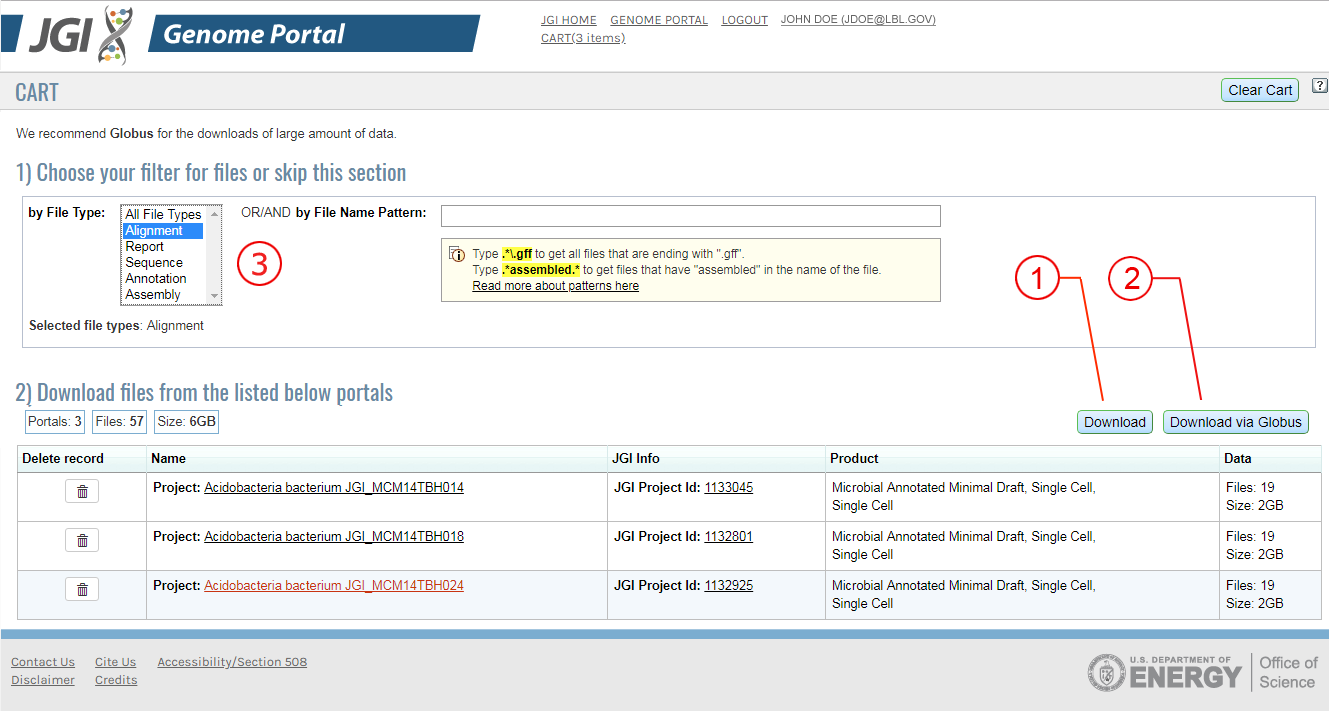
Filtering in the Cart (Figure B)
- Filter portals in the cart to limit the number or type of files you will download.
- Filter based on the file type (annotation, assembly, etc) or based on a file pattern.(Figure B, 3)
Download Files in Bulk(Figure B)
From the CART the users have 2 options to download.
- Download (Figure B, 1)
"Download" - prepares files on disk and notifies the user by email when the files are staged. Using this option will result in a download through your browser. Files are available on disk for 14 days
- Download via Globus (Figure B, 2)
"Download via Globus" process stages the files on disk (the user's Globus ID should be provided).The user is notified by email from JGI that the files are ready. Clicking on the link in email brings the Globus interface for data transfer from the JGI endpoint to the user's endpoint or local machine
Please provide your feedback
Download with web UI
To find the genome/metagenome/group of your interest please start from the home page. After narrowing down your results with filters ("Advanced Search" on the blue panel) please click on the link "Download" to proceed to download page of the particular portal/project/group.
For example, searching for "fusobacteria" on home page will give you ~100 results in the table. If you are interested in the taxonomic group "Fusobacteria" then click on the "Download" link in the "Resources" column or click on the name of the group and then go to "Download" section of this group. Please login in order to download.
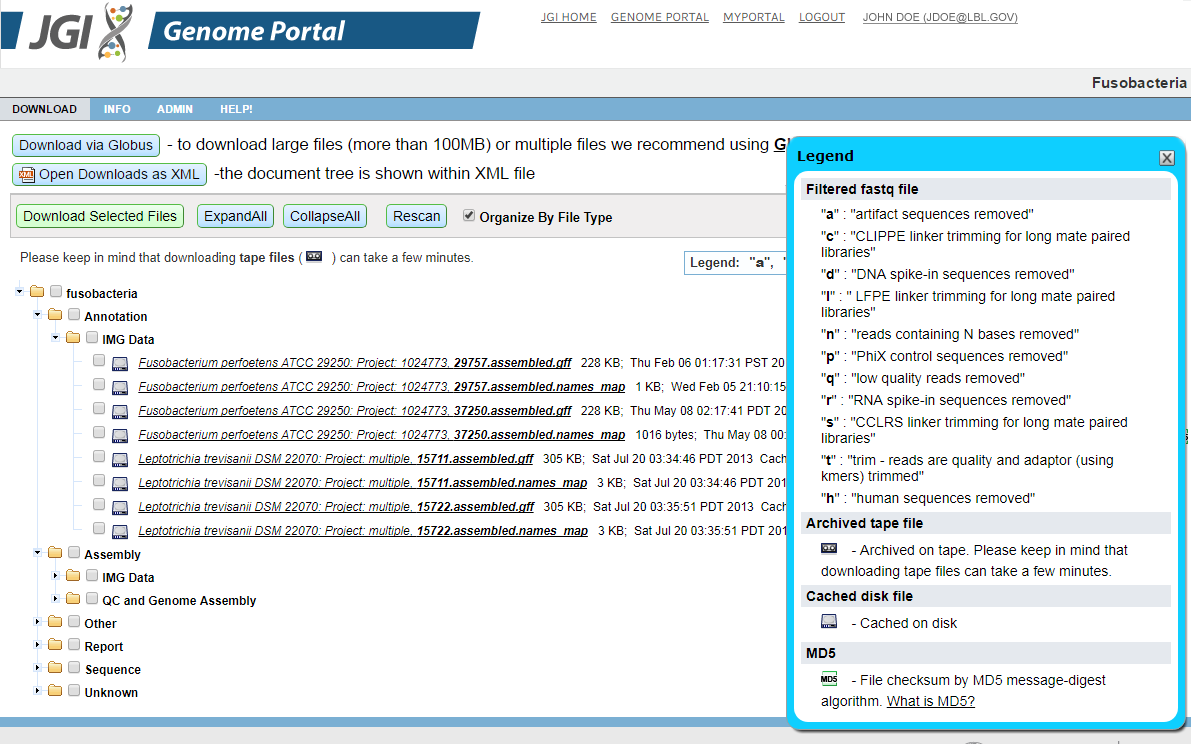
Downloads are available in a tree structure that divides the files into logical groups so that the user can download RAW files, assemblies, and so on with a single operation.
Available download files are presented in a tree based on the data-type (assembly, masked assembly, protein models, ESTs...). To expand or collapse all items in the tree, click Expand All or Collapse All. You can download files in one of two ways:
-
To download multiple files at once, select the checkboxes to the left of file sections or individual files, and hit the Download button. The selected items will be packaged into a .zip archive file and down-loaded to your browser. Note that you can select or deselect all download files by toggling the checkbox at the root of the tree.
-
To download an individual file, click on the desired single item in the tree.
New feature "Organize By File Type" for Downloads. You can use this option on the download page of Projects, Proposals and Groups. In order to use it please:
- Go to the download page
- Check the box "Organize By File Type".
- The folders on the "download tree" will be reorganized by the file type arrangement
Download with Globus service (fast, reliable, convenient)
How to set up your Globus account:
- Go to www.globus.org website.
- Create an account. Follow the instructions in email from Globus to activate your account.
- Globus transfers occur between the endpoints. The link to the JGI endpoint will be sent to you by email once you initiate the download. The other endpoint is your institution's Globus endpoint. To transfer to your local machine you can install the program Globus Connect Personal
How to use Globus services for your downloads
- Go to the download section of your project/portal/proposal/group
Please use search on the Home" page to locate the organism(s) of your interest.
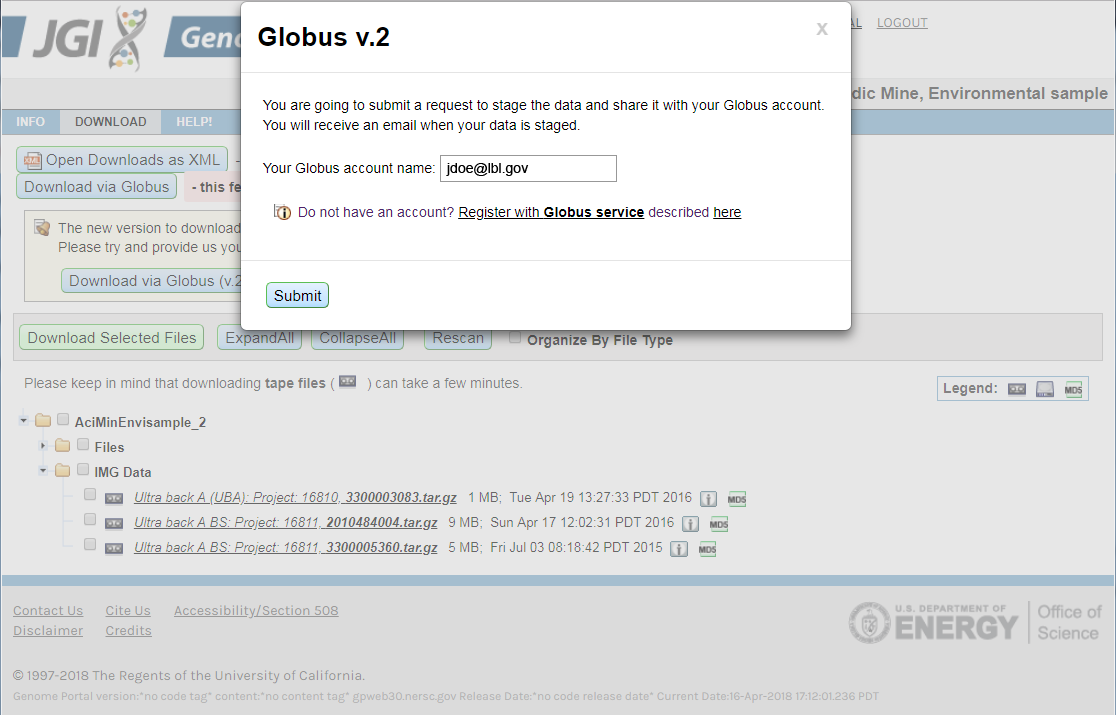
For example, for the proposal Acidic Mine, Environmental sample (Proposal ID: 2199 / 3483) start from the download page of this proposal portal. - Click the "Download via Globus (v.2)" button under the main navigation of the portal
- The dialog box will appear where you have to provide your Globus Account name.
- Submit the request and wait for the email from our service when your files are staged and ready for download.
NOTE: Please be aware that ALL files that belong to this portal will be staged on disk. Tape restore requests may be delayed until 8PM Pacific Time. - Click on the link in the received email.
- Globus then provides a user friendly interface for big data transfers.
- Provide your second endpoint you are planning to transfer to.
- Authenticate to your second endpoint.
- Select files and start to transfer.
- Behind the scenes the transfers are performed using GridFTP which is a parallel transfer protocol and program. GridFTP has built in checks that ensures the integrity of the transfers and guarantees that the files reach their destination intact.
Download with API
-
Identify the name of the portal before you can download.
You can find that using our JGI Portal search on the home page. Use any search terms necessary to find the portal you want, click on the "Download" link in the "Resources" column, then make a note of the short portal name in the URL. It is located between the second and third "/" characters in the path after the web host. For example, in the URL https://genome.jgi.doe.gov/portal/Aurpu_var_sub1/... the portal name to use for API download is "Aurpu_var_sub1"
You can also export the full search results into CSV format by clicking "Project Overview Report", then you could iterate over all your projects.The short portal name is identified in "Portal ID" column.
- Log in using the following command.
curl 'https://signon.jgi.doe.gov/signon/create' --data-urlencode 'login=USER_NAME' --data-urlencode 'password=USER_PASSWORD' -c cookies > /dev/null
Replace USER_NAME, USER_PASSWORD with the appropriate values
a) If you prefer to download directly from Portal, please follow the steps 3, 4 below
3. Download a list of files available for the portal that you are interested in.
For example, for PhytozomeV10 the command will look like this:
curl 'https://genome-downloads.jgi.doe.gov/portal/ext-api/downloads/get-directory?organism=PhytozomeV10' -b cookies > files.xml
4. Find the file that you would like to download in the XML document and download it. The details of XML schema (XSD) can be found here.
For example, if you look for "Alyrata_107_v1.0.annotation_info.txt", you will find the following entry in the file:
<file label="PhytozomeV10" filename="Alyrata_107_v1.0.annotation_info.txt" size="3 MB" sizeInBytes="3901148" timestamp="Sun Jan 12 17:46:56 PST 2014" url="/portal/ext-api/downloads/get_tape_file?blocking=true&url=/PhytozomeV10/download/_JAMO/53112a9e49607a1be0055980/Alyrata_107_v1.0.annotation_info.txt" project="" library="" md5="b03b5173b0adabe4c0e37f82b4a7a2a1"/>
Then you need to transfer the URL attribute from the entry to the download curl command (please make sure you that you replace "&" with "&").
The command to download the file would look like this:
curl 'https://genome-downloads.jgi.doe.gov/portal/ext-api/downloads/get_tape_file?blocking=true&url=/PhytozomeV10/download/_JAMO/53112a9e49607a1be0055980/Alyrata_107_v1.0.annotation_info.txt' -b cookies > Alyrata_107_v1.0.annotation_info.txt
b) OR to download with Globus, please follow these steps
3. Request the data to be staged for download via Globus.
For example, for PhytozomeV10 the command will look like this:
curl 'https://genome-downloads.jgi.doe.gov/portal/ext-api/downloads/globus/request' -b cookies --data-urlencode 'portal=PhytozomeV10' --data-urlencode '[email protected]'
Replace [email protected] with your Globus user name.
You can add the following optional parameters to the above curl command:
--data-urlencode 'fileTypes=type1,type2,...' (available types: Report, Alignment, Annotation, Assembly, Sequence)
--data-urlencode 'filePattern=<filename pattern>' (should be a Java regular expression, e.g. .*\.fasta\.gz)
--data-urlencode 'addedSince=YYYY-MM-DD' (if you are only interested in newer data)
--data-urlencode 'sendMail=true' (if you want to receive a notification via email when your data is ready)
--data-urlencode 'organizedByFileType=true' (if you want the data to be organized by file type)
4. Check the status of your request.
The curl command in step 3 will return a link that you can use to check the status of your request, for example:
curl 'https://genome-downloads.jgi.doe.gov/portal/ext-api/downloads/globus/NNNNN-MM/status' -b cookies
The command will return a link to your data when it is ready, for example:
Download request completed.
Data URL: https://app.globus.org/file-manager?origin_id=XXXXXX&origin_path=/NNNNN/MM/PhytozomeV10/&add_identity=UUUUUU
After that, you can either enter the URL in your Web browser or use the values of the "origin_id" and "origin_path" parameters of the URL with Globus API calls.
c) OR to download data in bulk, please follow these steps instead
3. Request multiple portals data to be staged.
curl 'https://genome-downloads.jgi.doe.gov/portal/ext-api/downloads/bulk/request' -b cookies --data-urlencode 'portals=portalName1,portalName2,...'
You can add the following optional parameters to the above curl command:
--data-urlencode 'fileTypes=type1,type2,...' (available types: Report, Alignment, Annotation, Assembly, Sequence)
--data-urlencode 'filePattern=<filename pattern>' (should be a Java regular expression, e.g. .*\.fasta\.gz)
--data-urlencode 'organizedByFileType=true' (if you want the data to be organized by file type)
--data-urlencode 'addedSince=YYYY-MM-DD' (if you are only interested in newer data)
--data-urlencode '[email protected]' (if you want to download the data via Globus)
--data-urlencode 'sendMail=true' (if you want to receive a notification via email when your data is ready)
4. Check the status of your request.
The curl command in step 3 will return a link that you can use to check the status of your request, for example:
curl 'https://genome-downloads.jgi.doe.gov/portal/ext-api/downloads/bulk/NNNNN-MM/status' -b cookies
The command will return a link to your data when it is ready, for example:
Download request completed.
Data URL: https://genome-downloads.jgi.doe.gov/portal/ext-api/downloads/bulk/NNNNN-MM/zip
or (if you added the "globusName" parameter to the data request):
Download request completed.
Data URL: https://app.globus.org/file-manager?origin_id=XXXXXX&origin_path=/NNNNN/MM/&add_identity=UUUUUU
IMG annotation files in "IMG Data" Folders
Please note that IMG annotation files are bundled in download_bundle.tar.gz
To learn about the contents of the tar bundle and how to extract them please read the IMG guidelines
Data Usage and Download Policy
For both new and old download pages, you are required to read and approve a JGI Data Usage Policy statement before accessing JGI data. This statement will appear on the first page you see when enter the download area, and may vary by organism. To continue to the download page, click the "Agree" button after reviewing the policy. You may also select the checkbox next to the "Agree" button to bypass the usage statement the next time you visit the download area for the given organism or group. If you would like to review the policy again please use "Show Data Usage Policy" button under the main navigation.