| |
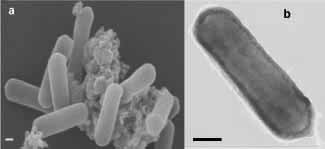
Electron micrographs of ‘N. maritimus’. a, Scanning
electron micrograph of Au/Pd sputtered cells. b, Transmission electron
micrograph of negative stained cells. Scale bars represent 0.1 μm.
|
For over a century, studies of aerobic ammonia-oxidizing microorganisms have been
limited to two narrow clades within the domain Bacteria [34, 1, 18, 26]. Despite the
application of molecular techniques to the characterization of natural microbial
communities and improved methodologies for cultivating microorganisms, general
understanding of the microbiota sustaining nitrification in soil and marine environments
has not been significantly modified until now. Our recent discovery of an autotrophic
ammonia-oxidizing archaeon [28], the first documentation of this physiology within the
domain Archaea, necessitates a reassessment of nitrifying diversity and of the potential
contribution of Archaea to global nitrogen and carbon cycles [28]. The isolate
(‘Nitrosopumilus maritimus’) belongs to the Crenarchaeota, one of the recognized
kingdoms in the domain Archaea. The general physiology of this isolate is
similar to that of characterized bacterial ammonia oxidizers, growing autotrophically via
the oxidation of ammonia to nitrite. It is the first representative of the
planktonic marine Crenarchaeota in pure culture. The marine Crenarchaeota, first
discovered in cultivation-independent molecular surveys over a decade ago [13, 20], may
represent some of the most abundant organisms on our planet. Estimated to number
approximately 1028 cells in the world’s oceans [23], they almost certainly play important
roles in global biogeochemical cycles.
The discovery of ‘N. maritimus’, by revealing significant gaps in our knowledge of
microbial ammonia oxidation, now raises a variety of critical questions about the
diversity, ecology, and evolutionary origins of ammonia oxidation-based metabolism. We
anticipate that the availability of a genome sequence will greatly accelerate studies of the
physiology of this novel isolate, offer an important perspective on the origin(s) and
diversification of ammonia-oxidizing microorganisms, and provide an essential
framework for interpreting partial genome sequence recovered from past and ongoing
environmental surveys of environments inhabited by low-temperature Crenarchaeota.
More generally, it should contribute to a better understanding of the biogeochemical
cycling of nitrogen and carbon.
References
1. Aakra, A., Utaker, J.B., Pommerening-Roser, A., Koops, H.P., and Nes, I.F.
(2001). Detailed phylogeny of ammonia-oxidizing bacteria determined by rDNA
sequences and DNA homology values. International Journal of Systematic and
Evolutionary Microbiology 51, 2021-30.
13. DeLong, E.F. (1992). Archaea in coastal marine environments. Proc Natl Acad
Sci U S A 89, 5685-9.
18. Egli, K., Langer, C., Siegrist, H.R., Zehnder, A.J.B., Wagner, M., and van der
Meer, J.R. (2003). Community analysis of ammonia and nitrite oxidizers during
start-up of nitritation reactors. Applied and Environmental Microbiology 69,
3213-22.
20. Fuhrman, J.A., McCallum, K., and Davis, A.A. (1992). Novel major
archaebacterial group from marine plankton. Nature 356, 148-9.
23. Karner, M.B., DeLong, E.F., and Karl, D.M. (2001). Archaeal dominance in the
mesopelagic zone of the Pacific Ocean. Nature 409, 507-10.
26. Koops, H.P., Purkhold, U., Pommerening-R?ser, A., Timmermann, G., and
Wagner, M. (2003). The lithotrophic ammonia-oxidizing bacteria, In The
Prokaryotes: An Evolving Electronic Resource for the Microbiological
Community, M. Dworkin, S. Falkow, E. Rosenberg, K.H. Schleifer, and E.
Stackebrandt, eds. (New York: Springer-Verlag), pp. http://link.springer-
ny.com/link/service/books/10125/.
28. K?nneke, M., Bernhard, A.E., de la Torre, J.R., Walker, C.B., Waterbury, J.B.,
and Stahl, D.A. (in press). Isolation of an autotrophic ammonia-oxidizing marine
archaeon. Nature.
34. Purkhold, U., Pommerening-Roser, A., Juretschko, S., Schmid, M.C., Koops,
H.P., and Wagner, M. (2000). Phylogeny of all recognized species of ammonia
oxidizers based on comparative 16S rRNA and amoA sequence analysis:
implications for molecular diversity surveyss. Appl Environ Microbiol 66, 5368-
82.
|

