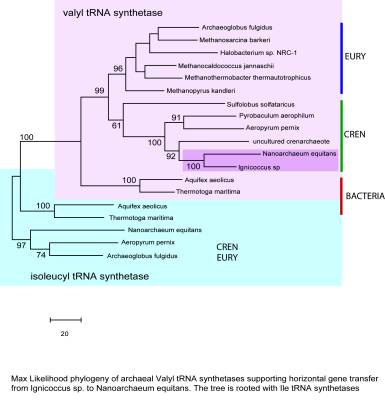
| A genomic perspective of the Nanoarchaeum equitans-Ignicoccus sp archaeal association Two years ago, the laboratory of Prof. Karl Stetter in Regensburg, Germany, reported the discovery of a new phylum of archaea, Nanoarchaeota (Huber et al., 2002). The organism that defined this new phylum, Nanoarchaeum equitans , was isolated from a submarine hydrothermal system of the coast of Iceland. What is remarkable about this hyperthermophile (optimum growth temperature ~90 degrees C, in the presence of S, H2 and CO2) is that it can only grow attached on another archaeum host, the crenarchaeote Ignicoccus sp KIN4/I. This and the extremely small size of N. equitans cells (~400 nm) suggested that this association represents a form of symbiosis or parasitism so far unique in archaea. The phylogenetic placement of Nanoarchaeota is deep, at the base of Archaea, suggesting that the phylum may be ancient. More recently, novel nanoarchaeotes have been detected by us and others in hyperthermophilic environments from other parts of the globe, including Yellowstone National Park, Kamchatka, China, Pacific hydrothermal vents, suggesting that these organisms may be quite abundant and diverse. A better understanding of the evolutionary history and the physiology of N. equitans and its host warranted a whole genome sequencing project. In collaboration between Dr. Stetter, Diversa and Celera, the genome of N. equitans was sequenced and published in 2003 (Waters et al, 2003). It was found that the genome is extremely small (0.49 Mbp) encoding 563 genes, with a relatively complete set of information and DNA repair genes but lacking entire pathways required for lipids, cofactors, aminoacids, nucleotides biosynthesis. This extreme genome reduction has been previously documented for obligate endosymbiotic orddd parasitic bacteria but is so far unique in Archaea. Similar to those cases, the N.equitans genome has drastically reduced the number of paralogous genes and has an extreme A/T bias (32%GC), which is remarkable considering it is a hyperthermophile. A unique feature of N. equitans is the presence of a relatively large number of split genes, which are transcribed as two fragments form different regions of the chromosome and assembled post-translationally into functional enzymes. The strict growth dependence on its host as well as the genomic proof of its metabolic incapability demonstrate that N. equitans has to acquire most of its biosynthetic precursors from Ignicoccus . Since Ignicoccus does not appear to suffer from the presence of sometimes more than a dozen N. equitans cells piled up on its membrane, it is likely that this specific association involves molecular recognition, signaling and exchange between the two archaea. Have any of the many metabolism genes missing in N. equitans been transferred to its host? Does the Ignicoccus genome include membrane transporters or protein subunits of pore complexes that could elucidate the mechanisms by which N. equitans aquires its building blocks and energy cofactors? These and other questions about the biology of this system would greatly benefit from knowing the complete genome sequence of the crenarchaeote Ignicoccus sp. KIN4/I. Because the N. equitans cells could not be separated from the Ignicoccus host, a mixed DNA library was used in the process of sequencing the N. equitans genome. Consequently, a relatively large fraction of the Ignicoccus genome has been sequenced during that project (~ 1Mb, encoding ~1200 genes, which, based on comparison with other similar archaea, represents over 50% of the genome). The Ignicoccus genome is 57%GC rich which eased the separate contig assembly for the two archaea. Having available such large sequence data of the host, I tried to answer some of the evolutionary questions related to the possible horizontal gene transfer between the two species. In N. equitans , which, based on rRNA and numerous protein genes appears to be closer to Euryarchaeota than to Crenarchaeota, there are a handful of genes that are crenarchaeal type, based on similarity values. Several of these genes, including those encoding valyl-tRNA synthetase, tyrosyl-tRNA synthetase, leucyl peptidase, translin, were available in the Ignicoccus sequence data, which allowed detailed phylogenetic analyses (example tree in figure below). Based on that, those genes appear to have been laterally transferred from Ignicoccus to Nanoarchaeum . This seems somewhat paradoxical, since the Nanoarchaeum genome is so much reduced, reflecting perhaps some strong selection pressure for maintaining certain genes aquired form its host and deleting some of its own. The transfer appear to have occurred a long time ago since the GC content in those transferred genes has been shifted to the N. equitans values. Based on the available Ignicoccus sequence data, no cases of lateral gene transfer from Nanoarchaeum were found so far. A complete picture of the extent of lateral gene transfer between these two species as well as the molecular basis of this unique archaeal association would require completion of the Ignicoccus genome sequence.
|
||
|
||
Ignicoccus hospitalis KIN4/I