| |
 |
Draft genome sequencing
of chlorinated-alkene-degrading
Polaromonas strain JS666
Basic facts about, importance of, and motivation for sequencing JS666:
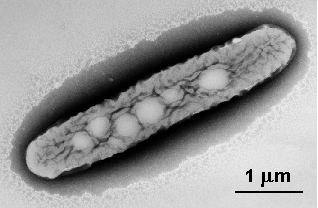
Polaromonas strain JS666 (ATCC No. BAA-500), a member
of the family Comamonadaceae in the beta-proteobacteria,
is a novel, aerobic, cis-dichloroethene (cDCE)-assimilating
organism with optimum growth at 20-25¾C (2). Strain JS666
is closely related to the Antarctic marine isolate Polaromonas
vacuolata (2). Significant phenotypic differences between
strain JS666 and Polaromonas vaculota exist. For example, Polaromonas
vaculota strains are white, motile, have gas vacuoles and
a temperature optimum around 4 degrees C (4). JS666, on the other
hand, is yellow, non-motile, devoid of vacuoles, and is not psychrophilic.
The substantial phylogenetic distance from other known aerobic
alkene-assimilating bacteria suggests a novel biochemistry for
cDCE oxidation.
Strain JS666 is the only aerobic organism known to use cDCE for energy
and growth. cDCE is a common groundwater contaminant (8) derived mainly
from incomplete anaerobic reductive dechlorination of the widely used chlorinated
solvents tetrachloroethene and trichloroethene (3, 6). The toxicity and
suspected carcinogenicity of cDCE qualifies it as an EPA priority pollutant,
and its presence in groundwater above concentrations of 70 ppb is considered
an unacceptable hazard to human health and the environment. Since growth-coupled
oxidation of cDCE does not appear to be common at field sites, JS666 is
a prime candidate for bioaugmentation at sites where cDCE has migrated
into aerobic zones.
In addition to the ability to degrade cDCE for growth, JS666 is capable
of transforming (though not growing upon) trans-1,2-dichloroethene
(tDCE), TCE, VC, 1,2-dichloroethane (1,2-DCA) and ETH (2). ETH is converted
to epoxyethane by cDCE-grown JS666 cultures, but not in succinate-grown
JS666 cultures, suggesting the a cDCE-inducible monooxygenase participates
in the cDCE pathway (2). Pulsed Field Gel Electrophoresis (PFGE) experiments
suggest that two large plasmids (approximately 340 and 360 kb) are present
in JS666. Further PFGE experiments suggest that the plasmids have a linear
topology. Additional work is required to determine if either of the plasmids
is associated with cDCE oxidation.
Recently, a Polaromonas was reported to be the organism responsible
for in situ biodegradation of naphthalene at a coal-tar-contaminated
site (5). Closely related strains have also been found recently in a variety
of contaminated sites (1, 7, 9), but their roles are unknown. The recent
isolation of the above strains suggests that members of the genus Polaromonas play
a major role in the subsurface degradation of environmental contaminants
that has been overlooked to date because of an emphasis on mesophilic bacteria.
References:
1. Alfreider, A., C. Vogt, and W. Babel. 2002. Microbial diversity in an
in situ reactor system treating monochlorobenzene contaminated groundwater
as revealed by 16S ribosomal DNA analysis. Syst. Appl. Microbiol. 25:232-240.
2. Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002.
Biodegradation of cis-dichloroethene as the sole carbon source
by a beta-proteobacterium. Appl. Environ. Microbiol. 68:2726-2730.
3. Distefano, T. D. 1999. The effect of tetrachloroethene on biological
dechlorination of vinyl chloride: Potential implication for natural bioattenuation.
Water Research 33:1688-1694.
4. Irgens, R. L., J. J. Gosink, and J. T. Staley. 1996. Polaromonas
vacuolata gen. nov., sp. nov., a psychrophilic, marine, gas vacuolate
bacterium from Antarctica. Int J Syst Bacteriol 46:822-6.
5. Jeon, C. O., W. Park, P. Padmanabhan, C. DeRito, J. R. Snape, and E.
L. Madsen. 2003. Discovery of a bacterium, with distinctive dioxygenase,
that is responsible for in situ biodegradation in contaminated sediment.
Proc Natl Acad Sci U S A 100:13591-13596.
6. Lorah, M. M., and L. D. Olsen. 1999. Degradation of 1,1,2,2-tetrachloroethane
in a freshwater tidal wetland: field and laboratory evidence. Environ.
Sci. Technol. 33:227-234.
7. Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-Mora, R.
Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S
rRNA to study the bacterial community of polychlorinated biphenyl-polluted
soil. Appl. Environ. Microbiol. 67:1874-1884.
8. Squillace, P. J., M. J. Moran, W. W. Lapham, C. V. Price, R. M. Clawges,
and J. S. Zogorski. 1999. Volatile organic compounds in untreated ambient
groundwater of the United States, 1985-1995. Environ. Sci. Technol. 33:4176-4187.
9. von Wintzingerode, F., B. Selent, W. Hegemann, and U. B. Gobel. 1999.
Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial
consortium. Appl. Environ. Microbiol. 65:283-286. |