| |
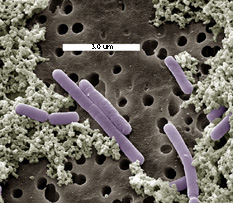
Photo courtesy of Jeff Broadbent,
Utah State University
|
Lactobacillus casei are
Gram-positive, facultatively anaerobic, non-motile and non-spore-forming,
rod-shaped (cell size range = 0.7-1.1 x 2.0-4.0 micrometer) members of
the industrially important lactic acid bacteria. Like other lactic acid
bacteria, L. casei are acid tolerant, cannot synthesize porphyrins,
and possess a strictly fermentative metabolism with lactic acid as the
major metabolic end product (Axelsson, 1998; Kandler and Weiss, 1986).
Within the genus Lactobacillus, L. casei form part of the
facultatively heterofermentative (“Group II”) species cluster,
which produce lactic acid from hexose sugars via the Embden-Meyerhof pathway
and from pentoses by the 6-phosphogluconate/phosphoketolase pathway (Axelsson,
1998). Growth of L. casei occurs at 15 but not 45oC, and requires
riboflavin, folic acid, calcium pantothenate, and niacin growth factors
(Kandler and Weiss, 1986).
L. casei are
a remarkably adaptive species, and may be isolated from raw and fermented
dairy products, fresh and fermented plant products, and the reproductive
and intestinal tracts of humans and other animals (Kandler and Weiss,
1986). Industrially, L. casei have application as human probiotics
(health-promoting live culture), as acid-producing starter cultures
for milk fermentation, and as specialty cultures for the intensification
and acceleration of flavor development in certain bacterial-ripened
cheese varieties (Fonden et al., 2000; Fox et al., 1998; Kosikowski,
1982).
The 9th edition
of Bergey’s Manual of Systematic Microbiology recognized four
subspecies of L. casei: L. casei subsp. casei, L.
casei subsp. pseudoplantarum, L. casei subsp. rhamnosus,
and L. casei subsp. tolerans (Kandler and Weiss, 1986).
However, more detailed phylogenetic studies have led to proposals that
members of the L. casei group be divided into three species: L.
rhamnosus, L. zeae and L. casei, with L. casei ATCC
334 as the neotype strain for the latter species (Chen et al., 2000;
Collins et al., 1989; Dellaglio et al., 1991; Dicks et al., 1996; Mori
et al., 1997). L. casei have a G + C content of 45-47%, and
the proposed neotype strain, ATCC 334, has a genome size previously
estimated at 2.2 Mbp (Ferrero et al., 1996; Kandler and Weiss, 1986;
Tynkkynen et al., 1999).
References:
- Axelsson, L.
1998. Lactic acid bacteria: classification and physiology, pp. 1-72.
In, S. Salminen and A. Von Wright (eds). Lactic Acid Bacteria: Microbiology
and Functional Aspects, 2nd ed. Marcel Dekker, Inc, New York.
- Chen, H., C.
K. lim, Y. K. Lee, and Y. N. Chan. 2000. Comparative analysis of
the genes encoding 23S-5S rRNA intergenic spacer regions of Lactobacillus
casei-related strains. Int. J. Syst. Evol. Microbiol. 50:471-478.
- Collins, M. D.,
B. A. Phyllips, and P. Zanoni. 1989. Deoxyribonucleic acid homology
studies of Lactobacillus casei, Lactobacillus paracasei sp. nov.,
subsp. paracasei and subsp. tolerans, and Lactobacillus
rhamnosus sp. nov., comb. nov. Int. J. Syst. Bacteriol.
39:105-108.
- Dellaglio, F.,
L. M. T. Dicks, M. du Toit, and S. Torriani. 1991. Designation of
ATCC334 in place of ATCC393 (NCDO 161) as the noetype straion of Lactobacillus
casei subsp. casei and rejection of the name Lactobacillus
paracasei (Collins et al., 1989). Int J. Syst. Bacteriol. 41:340-342.
- Dicks, L. M.
T., E. M. du Plessis, F. Dellaglio, and E. Lauer. 1996. Reclassification
of Lactobacillus casei subsp. casei ATCC 393 and Lactobacillus
rhamnosus ATCC15820 as Lactobacillus zeae nom. rev., designation
of ATCC 334 as the neotype of L. casei subsp. casei,
and rejection of the name Lactobacillus paracasei. Int. J.
Syst. Bacteriol. 46:337-340.
- Ferrero, M.,
C. Cesena, L. Morelli, G. Scolari, and M. Vescovo. 1996. Molecular
characterization of Lactobacillus casei strains. FEMS Microbiol.
Lett. 140:215-219.
- Fonden, R., G.
Mogensen, R. Tanaka, and S. Salminen. 2000. Effect of culture-containing
diary products on intestinal microflora, human nutrition and health – current
knowledge and future perspectives. International Dairy Federation
Bulletin number 352, IDF, Brussels.
- Fox, P.F., P.L.H.
McSweeney, and C.M. Lynch. 1998. Significance of non-starter lactic
acid bacteria in cheddar cheese. Aust. J. Dairy Technol. 53:83-89.
- Kandler, O.,
and N. Weiss. 1986. Genus Lactobacillus, pp. 1063-1065. In, P. H.
A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (eds.), Bergey's
Manual of Systematic Bacteriology, vol 2, 9th ed. Williams and Wilkins,
Baltimore.
- Kosikowski, F.V.
1982. Cheese and fermented milk foods, 2nd ed. Kosikowski and Assoc.,
Brooktondale, N.Y.
- Mori, K., K.
Yamazaki, T. Ishiyama, M. Katsumata, K. Kobayashi, Y. Kawai, N. Inoue,
and H. Shinano. 1997. Comparative sequence analyses of the genes
coding for 16S rRNA of Lactobacillus casei-related taxa. Int.
J. Syst. Bacteriol. 47:54-57.
- Tynkkynen, S.,
R. Satokari, M. Saarela, T. Mattila-Sandholm, and M. Saxelin. 1999.
Comparison of ribotyping, randomly amplified polymorphic DNA analysis,
and pulsed-field gel electrophoresis in typing of Lactobacillus
rhamnosus and L. casei strains. Appl. Environ. Microbiol.
65:3908-3914.
|

