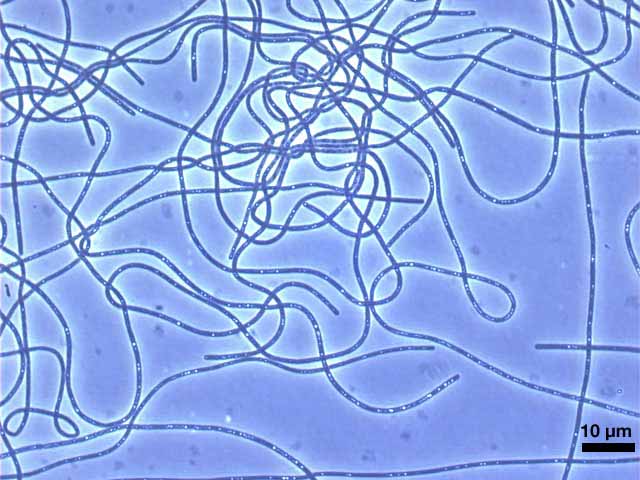
The phylum Chloroflexi is a deep-branching lineage within the Domain Bacteria. The members of the Chloroflexi are apparently metabolically diverse and widely distributed in nature. Although these organisms generally stain Gram-negative, a lipopolysaccharide-containing outer membrane is not present and the peptidoglycan is a variant that usually contains L-ornithine as the diamino acid. Most members of the Chloroflexi exhibit gliding motility. Members of the Family Chloroflexaceae are gliding filamentous, anoxygenic phototrophs. The “green” members of this family (Chloroflexus spp., Chloronema spp., Oscillochloris spp., and Chlorothrix spp.) synthesize bacteriochlorophylls a and c and use chlorosomes as their light harvesting antennae, while the “red” members of this family (Heliothrix and Roseiflexus spp.) only synthesize bacteriochlorophyll a and thus lack chlorosomes. Carbon dioxide fixation in these organisms occurs by the 3-hydroxypropionate pathway (Klatt et al., 2007). Chloroflexus aggregans was originally isolated from hot springs in Japan and was described by Hanada et al. (1995). The characteristics of this isolate are generally similar to those of other Chloroflexus spp. C. aggregans grew anaerobically as a photoheterotroph and aerobically as a chemoheterotrophy. The optimal growth temperature for C. aggregans is 55°C, and the mol% GC content of its DNA is 56.7%. The organism forms gliding filaments of variable length with a diameter of 1 to 1.5 µm. This organism forms mat-like aggregates in liquid medium that have the appearance of green-colored balls (Hanada et al., 2002). The name of the organisms comes from the observation that cellular aggregates reform rapidly whenever a growing culture is dispersed to form a uniform suspension. Physiological and biochemical studies suggest that this strain is distinct from Chloroflexus aurantiacus strains, and for this reason was given a different species designation. However, beyond these initial physiological studies, the properties of this organism are largely unknown. Characterization of the genome is anticipated to contribute significantly to our knowledge of the origins and evolution of photosynthesis and will help to understand anoxygenic phototrophic metabolism in significantly greater detail. References:
|
||
|
||
Chloroflexus aggregans DSM 9485