| |
Pseudomonas
fluorescens encompasses a group of common, nonpathogenic saprophytes
that colonize soil, water and plant surface environments. As its name implies,
it produces a soluble, greenish fluorescent pigment, particularly under
conditions of low iron availability. It is a obligate aerobe, except for
some strains that can utilize NO3 as an electron acceptor in place of O2.
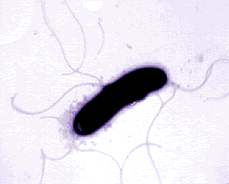
It is motile by means of multiple polar flagella. P. fluorescens has
simple nutritional requirements and grows well in mineral salts media supplemented
with any of a large number of carbon sources (1). Genetic techniques such
as conjugation, transposition, and gene replacement are well established.
Strain
Pf0-1 was isolated from agricultural soil and traits contribute to
its survival and growth in this environment are being studied (2,3).
Because they are well adapted to in soil, P. fluorescens strains
are being investigated extensively for use in applications that require
release and survival of bacteria in the soil. Chief among these are
bioremediation of various organic compounds, and biocontrol of pathogens
in agriculture.
Pseudomonads are
noted for their metabolic diversity and are often isolated from enrichments
designed to identify bacteria that degrade pollutants. Bioremediation
applications seek to exploit the inherent metabolic diversity of P.
fluorescens to partially or completely degrade pollutants such
as styrene, TNT and, polycyclic aromatic hydrocarbons (4-6) . In
addition, strains can be modified genetically to improve their performance
in particular applications.
A
number of strains of P. fluorescens suppress plant diseases
by protecting the seeds and roots from fungal infection (7). This
effect is the result of production of a number of secondary metabolites
including antibiotics, siderophores and hydrogen cyanide. Competitive
exclusion of pathogens as the result of rapid colonization of the
rhizosphere by P. fluorescens may also be an important factor
in disease control.
The
sequence of the P. fluorescens genome will provide insight
into its metabolic versatility and its interaction with the environment.
References
-
Palleroni,
N.J. (1984) Pseudomonadaceae. In Bergey's Manual of Systematic
Biology. Kreig, N.R., and Holt, J.G. (eds). Baltimore: The Williams
and Wilkins Co., pg. 141-199.
-
Compeau,
G., Al-Achi, B.J., Platsouka, E., and Levy, S.B. (1988) Survival
of rifampicin-resistant mutants of Pseudomonas fluorescens and
Pseudomonas putida in soil systems. Appl. Environ. Microbiol.
54: 2432-2438
-
DeFlaun,
M.F., Tanzer, A.S., McAteer, A.L., Marshall, B., and Levy, S.B.
(1990) Development and characterization of an adhesion-deficient
mutant of Pseudomonas fluorescens. Appl. Environ. Microbiol.
56: 112-119.
-
Baggi,
G., Boga, M.M., Catelani, D., Galli, E., and Treccani, V. (1983)
Styrene catabolism by a strain of Pseudomonas fluorescens. Syst.
Appl. Microbiol. 4: 141-147.
-
Gilcrease
P.C., and Murphy V.G. (1995) Conversion of 2,4-diamino-6-nitrotoluene
to a novel metabolite under anoxic and aerobic conditions. Appl
Environ Microbiol. 61: 4209-4214.
-
Caldini,
G., Cenci, G., Manenti, R., and Morozzi, G. (1995). The ability
of an environmental isolate of Pseudomonas fluorescens to utilize
chrysene and other four-ring polynuclear aromatic hydrocarbons.
Appl. Microbiol. Biotechnol. 44: 225-229.
-
O'
Sullivan, D.B., and O'Gara, F. (1992) Traits of fluorescent Pseudomonas
spp. involved in supression of plant root pathogens. Microbiol
Rev 56: 662-676.
|